Verbeterde workflow.
Gedeelde inzichten.
Ontdek onze bibliotheek met webinars
Bekijk eerder opgenomen webinars van deskundigen op het gebied van digitale en computergebaseerde pathologie
Waarom digitaal gaan?
Meer gevallen van kanker
Gezien het feit dat nieuwe gevallen van kanker in de komende twee decennia naar verwachting met bijna 70% zullen stijgen, krijgen pathologen steeds meer werk op hun bord...
Digitale pathologie
Digitale pathologie zorgt voor efficiëntere workflows en daarnaast biedt het pathologen de mogelijkheid om digitale beelden op allerlei verschillende en efficiënte manieren te gebruiken...
Digitale technologie versterkt uw expertise
Technologie stimuleert innovatie in de gezondheidszorg...
Toegenomen productiviteit
We willen de druk op pathologen verminderen door de worfklow te stroomlijnen...
Transformatie in pathologie


Ontdek ons Philips Digital & Computational Pathology-portfolio
Ontdek ons volledige productportfolio door hieronder te klikken:
Transformeer uw praktijk met digitale en computergebaseerde pathologie
Digitaliseer uw workflow
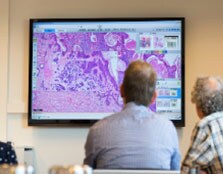
Zorg dat pathologen overal kunnen samenwerken

Een gestroomlijnde flow aan diagnostische informatie
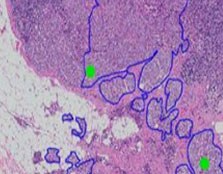
Help pathologen om sneller en preciezer een diagnose te stellen
Op zoek naar meer informatie?
U kunt meer informatie krijgen over digitale pathologie en computergebaseerde pathologie door u aan te melden voor updates. Blijf op de hoogte met de nieuwste artikelen, nieuws, recente opinies en meer:
“In difficult and diagnostically rare diseases, digital pathology will lead to expert diagnosis. And this at the end, will lead the best patient care”
Ivo Van Den Berghe
Head of Anatomical Pathology,
AZ-Sint-Jan, Bruges“Transitioning our entire workflow to digital processes demonstrates our commitment to ensuring our patients and clinical colleagues receive the fastest and best informed diagnoses possible”
Alexi Baidoshvili
Pathologist, LabPON“It is very important that IDEXX provides accurate diagnosis, getting the results through as quickly as possible”
Jenny McKay
Head of Anatomic Pathology, IDEXX Whetherby
Archief met webinars
Blijf met ons in contact

Volg ons op Twitter
Volg @Philips_Path en neem deel aan het gesprek en blijf op de hoogte van de nieuwste innovaties, nieuws en interessante feiten.

Neem contact met ons op via LinkedIn
Bij Philips willen we de levenskwaliteit van mensen verbeteren door middel van zinvolle innovatie. Kom met ons in contact om nooit meer iets te missen en deel te nemen aan het gesprek.

Bekijk onze nieuwste video's op YouTube
Ontdek hoe Philips innovatie biedt die belangrijk is voor u. Bekijk onze video's en zie hoe het vakgebied van de pathologie aan het veranderen is.
Philips IntelliSite Pathology Solution is in overeenstemming met Verordening (EU) 2017/746 van het Europees Parlement en de Raad van 5 april 2017 betreffende medische hulpmiddelen voor in-vitrodiagnostiek (IVDR).








