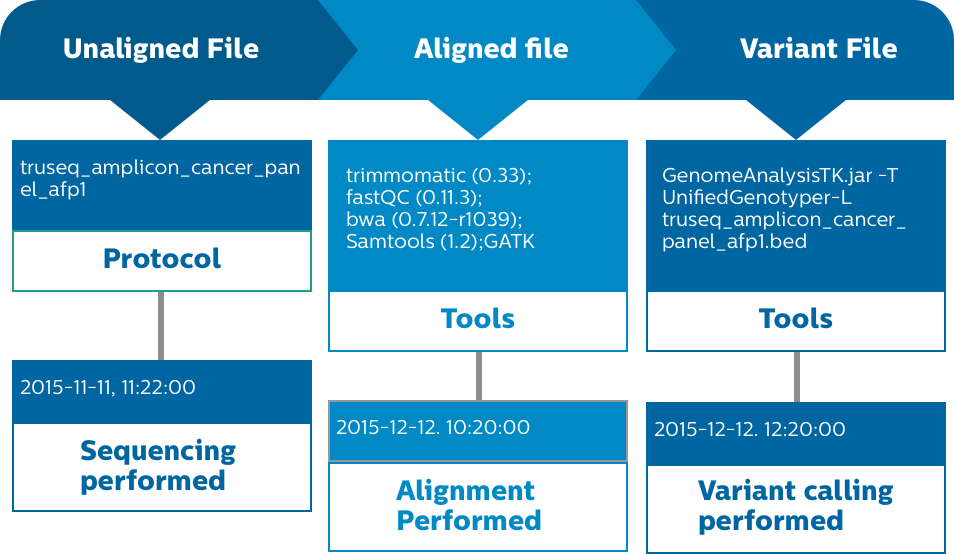
Processing of raw -omics data using automated pipelines with quality checks and traceability using Decode.
Our decode module is molecular test agnostic. We work with a variety of tests from sequencing by synthesis (SBS), semiconductor, and nanopore sequencing techniques to identifiy genomic abberations.
IntelliSpace Genomics offers full tracking and provenance of data and tools at each step
For each sample processed, all specific versions of your algorithms, parameters and databases used are recorded for complete traceability and reproducibility.
Extensibility to upload your own tools to customize and design pipelines
Open hardware compatibility gives healthcare organizations the flexibility to keep pace with rapidly evolving hardware technology.
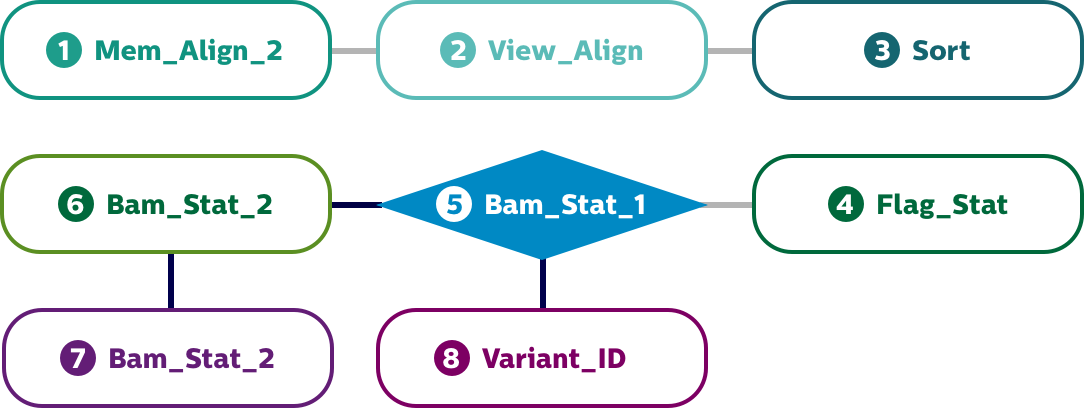
Benchmark your algorithms. We would be glad to do a proof of concept for you.
Want to learn more? Get in touch
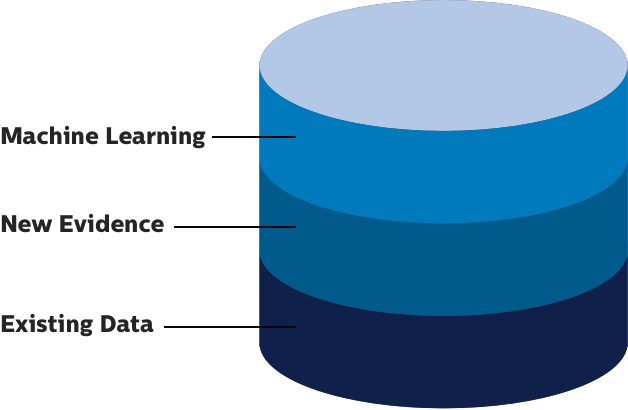
Create a knowledgebase that transforms previous data into new evidence with Curate.
Our customizable knowledgebases act as a collection of annotated associations between genomic aberrations and related phenotypes. Our machine learning leverages growing data.
Curated knowledgebase workflow
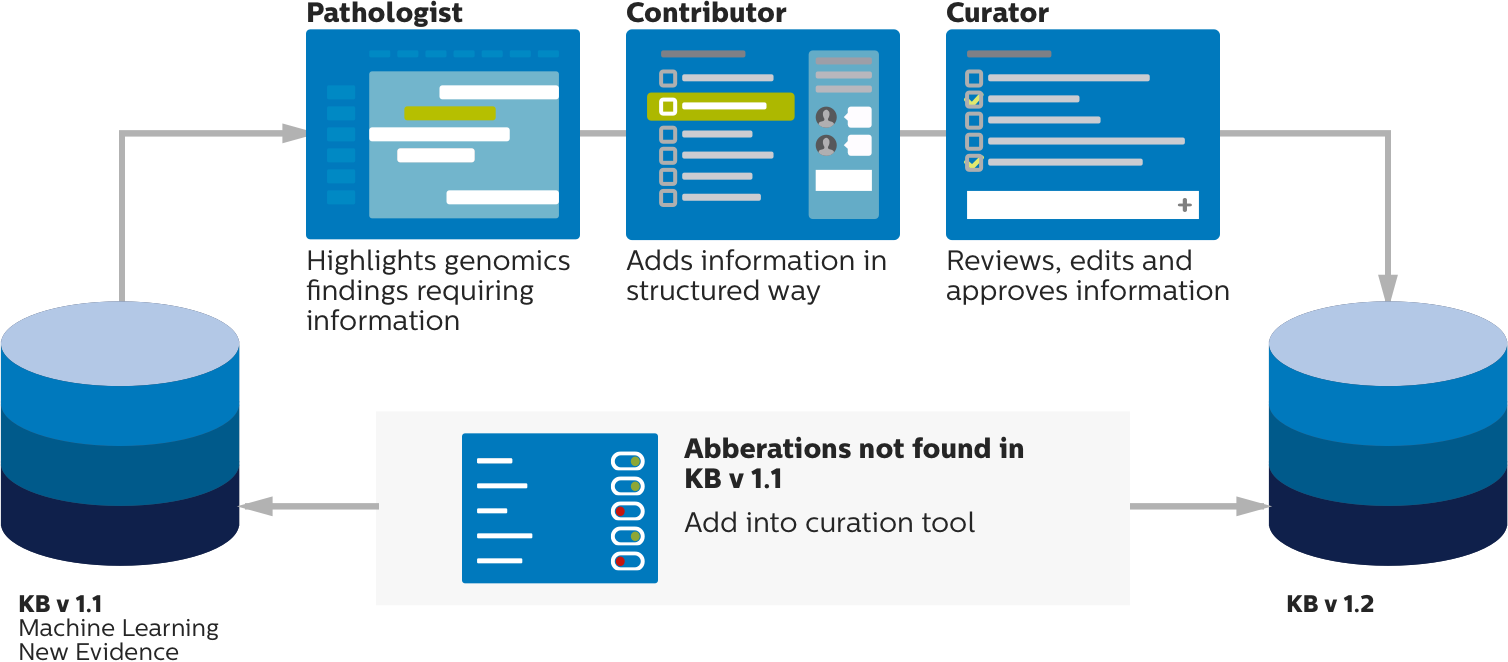
Seamless transition between the clinical workflow and curation workflow
Each entry in the knowledgebase is a documented genomic aberration association, which is relevant to understanding genomic drivers of disease and treatment response. Doctors and qualified personnel are allowed to curate this knowledgebase and to continue to enter observations.
Your curated data in a usable and scalable knowledge base.
The goal is to provide consistent and transparent clinical interpretation of patient genomic data for: 1. Improved diagnosis and treatment options 2. Targeted therapy development: research and discovery of variant associations for new therapeutic innovations
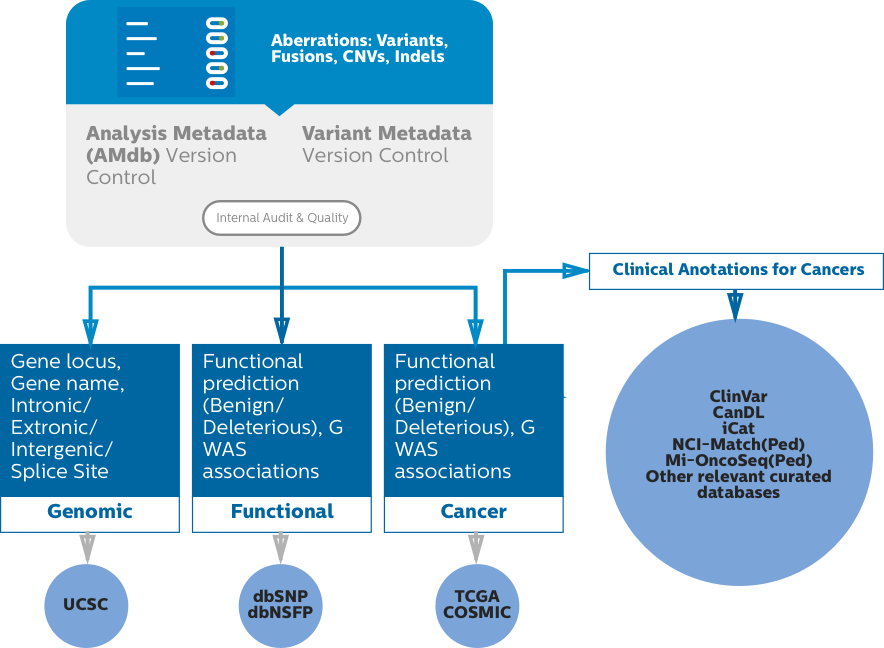
Variant filtering & annotation of processed genomic data according to functional, biological, pathway and disease indication with Annotate.
Variant Calling
Configurable prioritized list of variants from with information on location, functional annotation and QC information.
Variant Identification
Identify and verify variant detail against literature or your own knowledgebase.
Clinical Signature
Find variant-specific indications from literature references.
Surfacing exhaustive, up to date research and information on clinically relevant variants.
Our open annotation framework is a genomic knowledgebase that leverages comprehensive literature reviews and the latest clinical information.
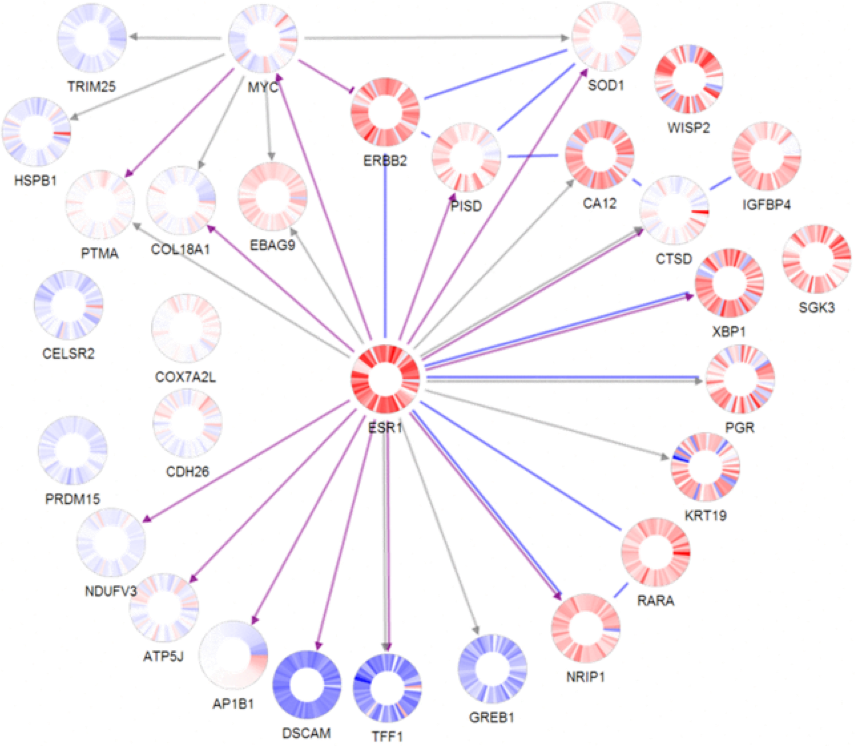
Dig deeper with powerful data visualization tools and deep research linking.
We give you the tools, data, and information to progressively discover more about the variants found in a patient’s genome. The example shown here is tumor pathway.
Identification of therapies and clinical trials based on patients' genotype and phenotype using Match.
Match offers clinicians molecular decision support by leveraging literature mining, clinical trial data, and selected knowledgebases.
Better matches
A pilot study using 80 clinical discharge summaries revealed that our ontology-driven semantic search framework effectively improves clinical task workflow and can contribute to the decision making process of the clinicians through faster, and more accurate information retrieval and visualization at the point of care.1
Unstructured data + Philips Clinical NLP = Better Matches, that get better with time.
1 Ontology-Driven Semantic Search for Brazilian Portuguese Clinical Notes. Stud Health Technol Inform. 2015;216:1022.
Light-up clinically actionable variants with filter
To speed up the reporting workflow, we uniquely embed the N-of-One variant database or your own curated knowledgebase natively on the platform. This integration enables lighting-up clinically actionable variants through filtering for rapid selection.
Highlight, annotate, filter, and prioritize
comprehensive genomic databases right inside IntelliSpace Genomics. This is in addition to all of the available public data sources the platform brings for comprehensive annotation, filtering and reliable prioritization.
Our unique integration with N-of-One brings together one of the most

What the integration with N-of-One means?
“Philips is a clear leader in clinical informatics, integrating and managing big data in the healthcare settings,” said Chris Cournoyer, CEO of N-of-One. “We believe that incorporating N-of-One’s Variant Database along with its patient case analysis and interpretation services will be further enabled by tight integration with this leading digital health solution.”
Intuitive filtering options
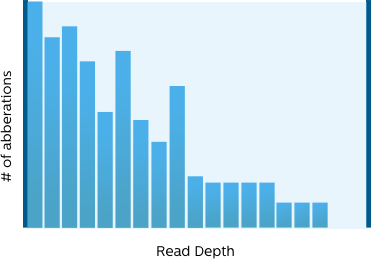
To focus on information and/or actionable variants from the large number detected by NGS, we provide flexible and visual filtering options. To make the filters easily accessible, we have classified them in QC, frequency, functional impact scores and database presence categories. Our visual filtering interface displays distribution of variants by read depth and allele frequency so that you can optimize your filters at a glance.